Graph-Dynamo: Learning stochastic cellular state transition dynamics from single cell data
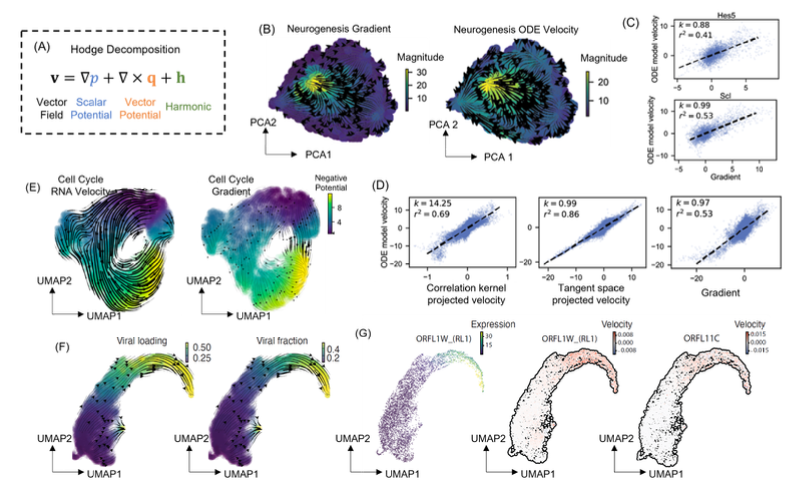
Modeling cellular processes in the framework of dynamical systems theories is a focused area in systems and mathematical biology, but a bottleneck to extend the efforts to genome-wide modeling is lack of quantitative data to constrain model parameters. With advances of single cell techniques, learning dynamical information from high throughput snapshot single cell data emerges as an exciting direction in single cell studies. Our previously developed dynamo framework reconstructs generally nonlinear genome-wide gene regulation relations from single cell expression state and either splicing- or metabolic labeling-based RNA velocity data. In this work, we first developed a graph-based machine learning procedure that imposes a mathematical constraint that the RNA velocity vectors lie in the tangent space of the low-dimensional manifold formed by the single cell expression data. Unlike a popular cosine correlation kernel used in literature, this tangent space projection (TSP) preserves the magnitude information of a vector when one transforms between different representations of the data manifold. Next, we formulated a data-driven graph Fokker-Planck (FPE) equation formalism that models the full cellular state transition dynamics as a convection-diffusion process on a data-formed graph network. The formalism is invariant under representation transformation and preserves the topological and dynamical properties of the system dynamics. Numerical tests on synthetic data and experimental scRNA-seq data demonstrate that the graph TSP/FPE formalism built from snapshot single cell data can recapitulate system dynamics.